Project 16. Understanding the influence of eRNA association modes on transcriptional condensates.
Multicellular organisms rely on differential gene expression to create cellular diversity. Precise gene regulation is crucial, as failures can lead to reduced fitness, developmental defects, and disease. Enhancers, DNA elements with binding sites for transcription factors, play a key role in this process. They regulate gene activation by coming into physical proximity of promoters and transferring some of the bound components. Recently, it has become clear that enhancer activity requires the formation of sub-micrometre-sized transcriptional condensates.
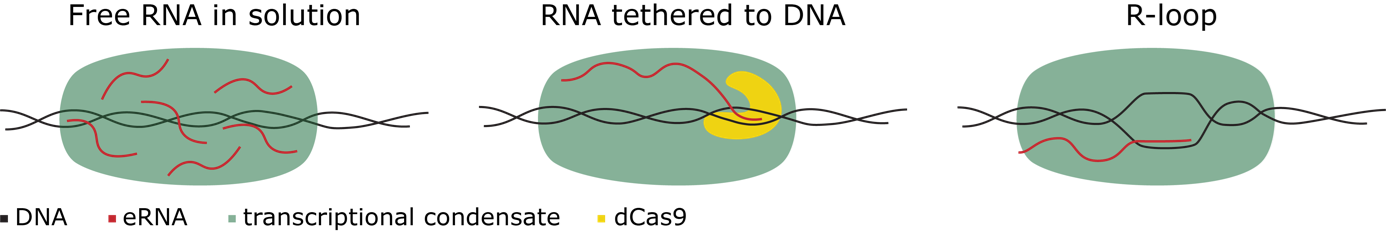
Enhancers also produce enhancer RNAs (eRNAs), and their transcriptional activity correlates with their ability to activate nearby genes. While proteins have been extensively studied in transcriptional condensate formation, the role of RNAs has received less attention. However, previous studies suggest that short RNAs like eRNAs can trigger phase separation of proteins in transcriptional condensates, and transcription factors can interact directly with RNAs. Given that most eRNAs only act locally (in cis), the presence of eRNAs at enhancer sites, often forming R-loop structures, may be more important than the specific eRNA molecule itself. This leads to the hypothesis that eRNAs play a crucial role in the formation and function of transcriptional condensates, and that the mode of their association with DNA (Figure 1) is a key determinant of condensate properties.
Because the impact of RNA on transcriptional condensate formation remains largely unexplored, this project aims to determine how eRNA association modes influence transcriptional condensate formation and properties. To this end, we will investigate three key aims:
- Impact on Transcription Factor Condensation: How does the mode of eRNA association (Figure 1) influence transcription factor condensation? What roles do eRNA properties (length, sequence, structure, number of molecules, modifications) play in this process?
- Formation of Surface Condensates: How does the mode of eRNA association affect surface condensate formation on single molecules of DNA? Can eRNAs trap transcription factors at the DNA through direct interactions?
- Influence on DNA:DNA Interaction: How do different eRNA association modes impact DNA:DNA contacts between enhancers and promoters? What are the effects of RNA properties on the strength and nature of these interactions?
To investigate these questions, we will utilize cutting-edge single-molecule assays and bulk phase separation techniques to reconstitute transcriptional condensate in vitro. This allows us to precisely control eRNA association modes – freely diffusing, tethered via dCas9, or anchored as R-loops – and observe their effects on condensate formation and function. We will also leverage collaborations to improve our single-molecule assay (Anand), to investigate the role of specific proteins (Dormann), and to test enzymatic activity (Padeken) in regulating eRNA association and condensate dynamics.
This project will provide fundamental insights into the mechanisms governing transcriptional condensate formation and function. By understanding how eRNA association mode influences these processes, we can gain a deeper understanding of gene regulation and its implications for development and disease.
